@Feelsbadman.jpg I searched and found references to 5ar contributing to insulin resistance but not the other way around...?
Among the top significant TFs associated with transcriptional regulation network (Supplementary Table 7) SP1, ESR1, GCR-alpha (NR3C1) and FOS were included in the Androgenetic Alopecia accepted. On the other hand, CREB1, c-Myc (MYC), Androgen receptor (AR), p53 (TP53), c-Jun (JUN), Oct ¾, RelA (RELA) and YY1 were among a set of TFs that are not found directly in the
dataset, but significantly connected to several objects in it (“hidden”).
@InBeforeTheCure, you not surprised with the proto-oncogenes coming up again?
http://www.sciencedirect.com/science/article/pii/S0040816616301720
c-Myc activity induces epidermal and sebocyte differentiation in HFSCs at the expense of hair follicle differentiation (see for example
Arnold and Watt, 2001, and
Frye et al., who suggest that c-Myc activation may promote epidermal/sebocyte differentiation by decreasing expression of focal adhesion related genes). The interesting thing in the paper you've posted is that beta-catenin apparently induces c-Myc and promotes epidermal differentiation, but beta-catenin is generally pro-HF differentiation so it's a bit surprising.
There was also
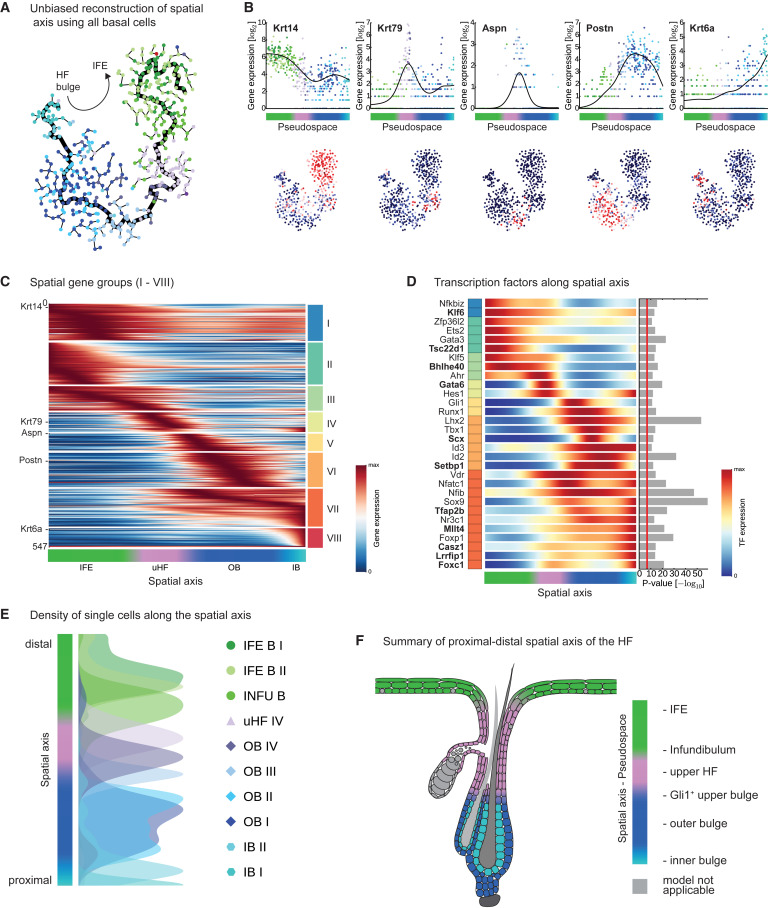
this transcriptomic study on different populations of stem cells in telogen epidermis in mice. I ran the signature genes for each spatial cluster through TFacts, and it found that one of the epidermal clusters (Cluster I) was highly enriched for c-Myc target genes:
Cluster I - MYC (p = 0, e = 0)
Cluster II - GLI2 (p = 0, e = 0), CTNNB1 (p = 4.4e-4, e = 1.76e-2), TP53 (p = 5.6e-4, e = 2.24e-2)
Cluster III - GLI1 (p = 4.9e-4, e = 1.764e-2)
Cluster IV - CTNNB1 (p = 5.1e-4, e = 1.683e-2)
Cluster V - USF2 (p = 5e-4, e = 1.7e-2), USF1 (p = 9.8e-4, e = 3.332e-2)
Cluster VI - SMAD5 (p = 1.4e-4, e = 5.18e-3), CTNNB1 (p = 8.3e-4, e = 3.071e-2)
Cluster VII - CTNNB1 (p = 0, e = 0), LEF1 (p = 5e-5, e = 2.1e-3)
Cluster VIII - SP1 (p = 3.2e-4, e = 9.28e-3), CTNNB1 (p = 1.02e-3, e = 2.958e-2)
Proto-oncogenes and tumor suppressors by nature are involved in cell proliferation, differentiation, apoptosis, senescence, and so on so it's not really surprising that they would be involved when there are known alterations to these processes in A.G.A.
Regarding common baldness the Hamilton's studies are like Mendel's with colors and flowers
New insight in this issue
http://www.nature.com/articles/ncomms14694
Meta-analysis identifies novel risk loci and yields systematic insights into the biology of male-pattern baldness
Yeah, a lot of the gene regions identified in that study were also found by Hagenaars et al. The latter found more regions due to larger sample size, but there are still some new ones in this meta-analysis. After combining them, I think that makes ~115 genetic regions associated with A.G.A so far.
 .
.