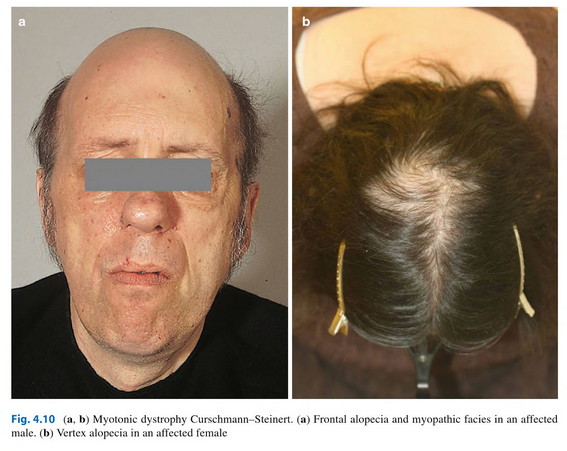
- Reaction score
- 950
I just stumbled upon this Biorxiv preprint of a new GWAS run on a sample of over 50,000 men.
Abstract (http://biorxiv.org/content/early/2016/08/31/072306):
Full text: http://biorxiv.org/content/biorxiv/early/2016/08/31/072306.full.pdf
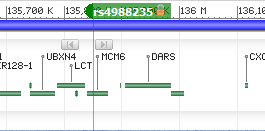
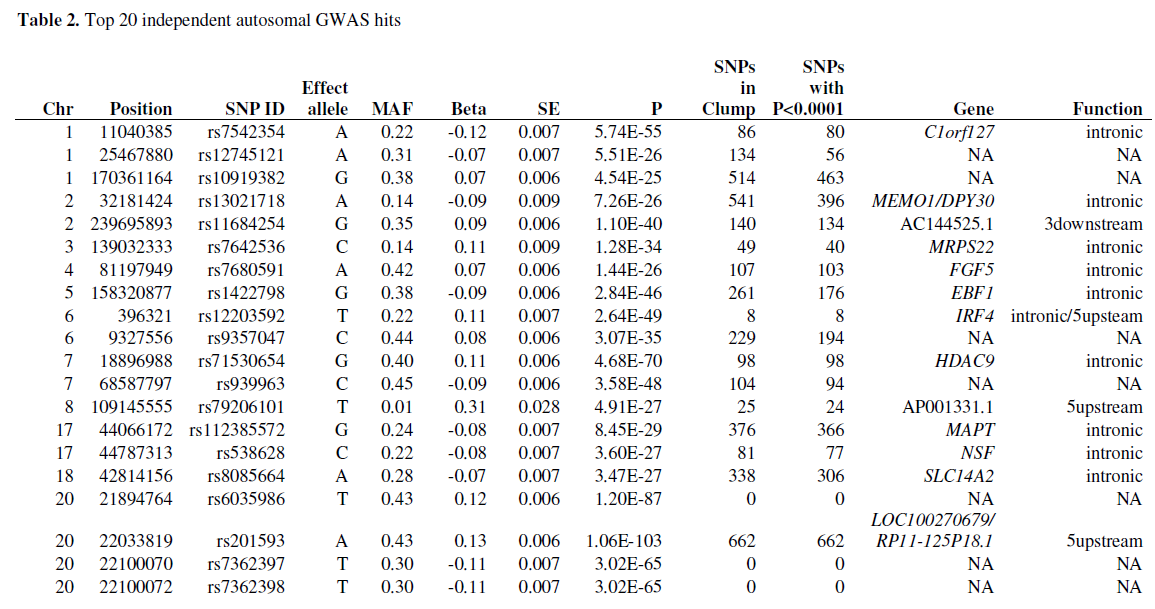
Table of Top 20 autosomal GWAS hits:
List of 112 associated autosomal genes:
List of 13 X-chromosome genes:
Some discussion from the paper:
Seems simple, right?
Abstract (http://biorxiv.org/content/early/2016/08/31/072306):
Male pattern baldness can have substantial psychosocial effects, and it has been phenotypically linked to adverse health outcomes such as prostate cancer and cardiovascular disease. We explored the genetic architecture of the trait using data from over 52,000 male participants of UK Biobank, aged 40-69 years. We identified over 250 independent novel genetic loci associated with severe hair loss. By developing a prediction algorithm based entirely on common genetic variants, and applying it to an independent sample, we could discriminate accurately (AUC = 0.82) between those with no hair loss from those with severe hair loss. The results of this study might help identify those at the greatest risk of hair loss and also potential genetic targets for intervention.
Full text: http://biorxiv.org/content/biorxiv/early/2016/08/31/072306.full.pdf
Table of Top 20 autosomal GWAS hits:

List of 112 associated autosomal genes:
GENE CHR START STOP NSNPS P
RSPO2 8 108911544 109095913 559 4.21E-23
EBF1 5 158122923 158526788 976 8.50E-22
PRR23B 3 138737873 138739768 1 3.03E-18
FAM53B 10 126307863 126432930 322 2.15E-17
NSF 17 44668035 44834830 90 2.34E-15
PDGFA 7 536895 559481 133 2.43E-15
WNT10A 2 219745255 219758651 22 3.14E-15
TBX15 1 119425666 119532179 327 3.39E-15
CENPW 6 126661253 126669754 14 3.48E-15
PLEKHM1 17 43513266 43568146 73 3.52E-15
GORAB 1 170501263 170522974 47 6.90E-15
FGF5 4 81187742 81212171 91 9.13E-15
KLF15 3 126061478 126076236 28 9.34E-15
CRHR1 17 43697710 43913194 362 1.99E-14
WNT3 17 44841686 44896082 107 2.20E-14
RUNX3 1 25226002 25291501 183 3.33E-14
C10orf11 10 77542519 78317130 2218 3.51E-14
TWIST2 2 239756673 239795893 159 4.47E-14
LOC100287387 2 239755218 239756320 3 6.97E-14
PRDM6 5 122424841 122523745 464 9.91E-14
OFCC1 6 9607550 9977826 1358 3.90E-13
MAPT 17 43971748 44105700 129 4.58E-13
METTL10 10 126447406 126480439 81 5.45E-13
WARS2 1 119573839 119683295 300 1.03E-12
SETBP1 18 42260138 42648475 973 2.42E-12
SLC14A2 18 42792947 43263072 1865 6.12E-12
KANSL1 17 44107282 44302740 179 1.35E-11
TOP1 20 39657462 39753127 196 1.74E-11
AUTS2 7 69063905 70257885 3099 4.81E-11
PRR23A 3 138722804 138725110 8 5.04E-11
DKK2 4 107842959 107957453 352 8.11E-11
BRE 2 28113482 28561768 1302 9.91E-11
FAF1 1 50906935 51425936 990 1.22E-10
C1orf127 1 11006530 11042094 141 1.24E-10
TRPS1 8 116420724 116681228 533 1.45E-10
YIPF4 2 32502958 32531658 84 1.54E-10
WNT6 2 219724546 219738954 22 2.09E-10
IRF4 6 391739 411443 82 2.28E-10
LGR4 11 27387508 27494334 247 3.18E-10
PLCG1 20 39766161 39804357 77 4.44E-10
PTHLH 12 28111017 28124916 53 5.79E-10
TRMT11 6 126307576 126360422 60 5.80E-10
TARDBP 1 11072679 11085549 27 5.83E-10
HINT3 6 126277861 126301390 42 6.22E-10
GFOD2 16 67708436 67753273 84 1.06E-09
MEMO1 2 32092892 32235698 373 1.53E-09
FADS2 11 61595713 61634825 140 1.68E-09
FADS1 11 61567097 61584529 30 1.79E-09
SSPN 12 26348269 26387710 127 2.15E-09
THADA 2 43457975 43823185 1227 2.45E-09
IQGAP1 15 90931473 91045475 374 2.84E-09
ATG7 3 11314010 11599139 828 3.51E-09
CLIC4 1 25071760 25170815 141 3.77E-09
RASA2 3 141205926 141331197 217 6.79E-09
MASP2 1 11086580 11107296 75 8.43E-09
ZHX3 20 39807089 39928739 285 9.61E-09
EIF3E 8 109213972 109260959 167 1.09E-08
SLC16A10 6 111408781 111544608 308 1.48E-08
CASZ1 1 10696661 10856707 553 1.54E-08
ATP6V0D1 16 67471917 67515089 71 1.75E-08
HDAC9 7 18126572 19036993 2945 1.79E-08
PAX3 2 223064606 223163715 362 1.90E-08
SRD5A2 2 31749656 31806040 178 1.90E-08
TSNAXIP1 16 67841010 67861971 38 2.39E-08
ZBTB38 3 141043055 141168634 336 2.39E-08
ACBD4 17 43209967 43221543 24 2.75E-08
SLC12A4 16 67977377 68002597 41 4.23E-08
PEX14 1 10535003 10690815 475 5.47E-08
EPB41L2 6 131160487 131384462 764 5.59E-08
XDH 2 31557188 31637611 324 6.62E-08
TMEM91 19 41869871 41889988 52 7.96E-08
PRDM4 12 108126643 108154914 89 8.53E-08
ZDHHC1 16 67428322 67450339 43 8.96E-08
SUCNR1 3 151591431 151599876 30 9.96E-08
PAX1 20 21686297 21696620 13 1.19E-07
ITCH 20 32951062 33099197 326 1.41E-07
B9D2 19 41860322 41870078 29 1.47E-07
VPS9D1 16 89773541 89787394 51 1.79E-07
C4orf22 4 81256874 81884910 2226 2.07E-07
RANBP10 16 67757005 67840555 153 2.21E-07
PWP1 12 108079590 108106257 90 2.40E-07
NUTF2 16 67880819 67905219 47 2.69E-07
PRKAG3 2 219687106 219696512 16 4.07E-07
HSD11B2 16 67465036 67471456 12 4.15E-07
TTC27 2 32853087 33046118 847 5.55E-07
RGS22 8 100973276 101118344 339 5.59E-07
CTCF 16 67596310 67673088 146 5.77E-07
METTL9 16 21610856 21668792 107 6.12E-07
NOP58 2 203130515 203168384 89 6.17E-07
TCF12 15 57210833 57580716 1181 6.34E-07
TMEM258 11 61556602 61560085 7 6.40E-07
LRP6 12 12268959 12419811 403 6.58E-07
ZNF462 9 109625378 109773807 331 7.61E-07
LRRC36 16 67360747 67419109 130 8.43E-07
SPAG17 1 118496288 118727848 528 1.04E-06
TCHH 1 152078793 152086556 16 1.05E-06
KIAA2012 2 202937978 202978327 147 1.14E-06
SUPT3H 6 44796469 45345670 1985 1.48E-06
CTNNB1 3 41240942 41281939 75 1.49E-06
PCGF3 4 699573 764428 328 1.83E-06
PLA2G15 16 68279247 68294961 38 1.86E-06
ELAVL4 1 50513686 50667540 294 1.90E-06
FAM175B 10 126490354 126525239 75 1.92E-06
ZNRF3 22 29279755 29453476 510 1.93E-06
FAM169B 15 98980391 99057611 281 2.17E-06
CENPT 16 67862060 67881361 33 2.24E-06
FYN 6 111982479 112194655 722 2.26E-06
ATP5SL 19 41937223 41945843 29 2.42E-06
REV3L 6 111620234 111804414 463 2.66E-06
MYRF 11 61520121 61555989 76 2.69E-06
RALY 20 32581732 32668067 157 2.70E-06
DLG5 10 79550549 79686348 492 2.73E-06
RSPO2 8 108911544 109095913 559 4.21E-23
EBF1 5 158122923 158526788 976 8.50E-22
PRR23B 3 138737873 138739768 1 3.03E-18
FAM53B 10 126307863 126432930 322 2.15E-17
NSF 17 44668035 44834830 90 2.34E-15
PDGFA 7 536895 559481 133 2.43E-15
WNT10A 2 219745255 219758651 22 3.14E-15
TBX15 1 119425666 119532179 327 3.39E-15
CENPW 6 126661253 126669754 14 3.48E-15
PLEKHM1 17 43513266 43568146 73 3.52E-15
GORAB 1 170501263 170522974 47 6.90E-15
FGF5 4 81187742 81212171 91 9.13E-15
KLF15 3 126061478 126076236 28 9.34E-15
CRHR1 17 43697710 43913194 362 1.99E-14
WNT3 17 44841686 44896082 107 2.20E-14
RUNX3 1 25226002 25291501 183 3.33E-14
C10orf11 10 77542519 78317130 2218 3.51E-14
TWIST2 2 239756673 239795893 159 4.47E-14
LOC100287387 2 239755218 239756320 3 6.97E-14
PRDM6 5 122424841 122523745 464 9.91E-14
OFCC1 6 9607550 9977826 1358 3.90E-13
MAPT 17 43971748 44105700 129 4.58E-13
METTL10 10 126447406 126480439 81 5.45E-13
WARS2 1 119573839 119683295 300 1.03E-12
SETBP1 18 42260138 42648475 973 2.42E-12
SLC14A2 18 42792947 43263072 1865 6.12E-12
KANSL1 17 44107282 44302740 179 1.35E-11
TOP1 20 39657462 39753127 196 1.74E-11
AUTS2 7 69063905 70257885 3099 4.81E-11
PRR23A 3 138722804 138725110 8 5.04E-11
DKK2 4 107842959 107957453 352 8.11E-11
BRE 2 28113482 28561768 1302 9.91E-11
FAF1 1 50906935 51425936 990 1.22E-10
C1orf127 1 11006530 11042094 141 1.24E-10
TRPS1 8 116420724 116681228 533 1.45E-10
YIPF4 2 32502958 32531658 84 1.54E-10
WNT6 2 219724546 219738954 22 2.09E-10
IRF4 6 391739 411443 82 2.28E-10
LGR4 11 27387508 27494334 247 3.18E-10
PLCG1 20 39766161 39804357 77 4.44E-10
PTHLH 12 28111017 28124916 53 5.79E-10
TRMT11 6 126307576 126360422 60 5.80E-10
TARDBP 1 11072679 11085549 27 5.83E-10
HINT3 6 126277861 126301390 42 6.22E-10
GFOD2 16 67708436 67753273 84 1.06E-09
MEMO1 2 32092892 32235698 373 1.53E-09
FADS2 11 61595713 61634825 140 1.68E-09
FADS1 11 61567097 61584529 30 1.79E-09
SSPN 12 26348269 26387710 127 2.15E-09
THADA 2 43457975 43823185 1227 2.45E-09
IQGAP1 15 90931473 91045475 374 2.84E-09
ATG7 3 11314010 11599139 828 3.51E-09
CLIC4 1 25071760 25170815 141 3.77E-09
RASA2 3 141205926 141331197 217 6.79E-09
MASP2 1 11086580 11107296 75 8.43E-09
ZHX3 20 39807089 39928739 285 9.61E-09
EIF3E 8 109213972 109260959 167 1.09E-08
SLC16A10 6 111408781 111544608 308 1.48E-08
CASZ1 1 10696661 10856707 553 1.54E-08
ATP6V0D1 16 67471917 67515089 71 1.75E-08
HDAC9 7 18126572 19036993 2945 1.79E-08
PAX3 2 223064606 223163715 362 1.90E-08
SRD5A2 2 31749656 31806040 178 1.90E-08
TSNAXIP1 16 67841010 67861971 38 2.39E-08
ZBTB38 3 141043055 141168634 336 2.39E-08
ACBD4 17 43209967 43221543 24 2.75E-08
SLC12A4 16 67977377 68002597 41 4.23E-08
PEX14 1 10535003 10690815 475 5.47E-08
EPB41L2 6 131160487 131384462 764 5.59E-08
XDH 2 31557188 31637611 324 6.62E-08
TMEM91 19 41869871 41889988 52 7.96E-08
PRDM4 12 108126643 108154914 89 8.53E-08
ZDHHC1 16 67428322 67450339 43 8.96E-08
SUCNR1 3 151591431 151599876 30 9.96E-08
PAX1 20 21686297 21696620 13 1.19E-07
ITCH 20 32951062 33099197 326 1.41E-07
B9D2 19 41860322 41870078 29 1.47E-07
VPS9D1 16 89773541 89787394 51 1.79E-07
C4orf22 4 81256874 81884910 2226 2.07E-07
RANBP10 16 67757005 67840555 153 2.21E-07
PWP1 12 108079590 108106257 90 2.40E-07
NUTF2 16 67880819 67905219 47 2.69E-07
PRKAG3 2 219687106 219696512 16 4.07E-07
HSD11B2 16 67465036 67471456 12 4.15E-07
TTC27 2 32853087 33046118 847 5.55E-07
RGS22 8 100973276 101118344 339 5.59E-07
CTCF 16 67596310 67673088 146 5.77E-07
METTL9 16 21610856 21668792 107 6.12E-07
NOP58 2 203130515 203168384 89 6.17E-07
TCF12 15 57210833 57580716 1181 6.34E-07
TMEM258 11 61556602 61560085 7 6.40E-07
LRP6 12 12268959 12419811 403 6.58E-07
ZNF462 9 109625378 109773807 331 7.61E-07
LRRC36 16 67360747 67419109 130 8.43E-07
SPAG17 1 118496288 118727848 528 1.04E-06
TCHH 1 152078793 152086556 16 1.05E-06
KIAA2012 2 202937978 202978327 147 1.14E-06
SUPT3H 6 44796469 45345670 1985 1.48E-06
CTNNB1 3 41240942 41281939 75 1.49E-06
PCGF3 4 699573 764428 328 1.83E-06
PLA2G15 16 68279247 68294961 38 1.86E-06
ELAVL4 1 50513686 50667540 294 1.90E-06
FAM175B 10 126490354 126525239 75 1.92E-06
ZNRF3 22 29279755 29453476 510 1.93E-06
FAM169B 15 98980391 99057611 281 2.17E-06
CENPT 16 67862060 67881361 33 2.24E-06
FYN 6 111982479 112194655 722 2.26E-06
ATP5SL 19 41937223 41945843 29 2.42E-06
REV3L 6 111620234 111804414 463 2.66E-06
MYRF 11 61520121 61555989 76 2.69E-06
RALY 20 32581732 32668067 157 2.70E-06
DLG5 10 79550549 79686348 492 2.73E-06
List of 13 X-chromosome genes:
GENE CHR START STOP NSNPS P
ITIH6 X 54775332 54824673 4 5.67E-05
KLF8 X 56258822 56314322 2 2.53E-08
FAAH2 X 57313110 57515629 8 9.96E-13
ARHGEF9 X 62854848 63005426 7 1.11E-11
MTMR8 X 63487961 63615333 4 1.12E-06
ZC4H2 X 64136250 64254593 3 5.32E-10
VSIG4 X 65241580 65259967 3 1.09E-35
HEPH X 65382433 65487231 5 1.16E-96
EDA2R X 65815479 65859140 2 5.13E-190
AR X 66763874 66944119 6 2.02E-269
OPHN1 X 67262186 67653299 41 4.25E-190
STARD8 X 67867511 67945684 12 1.56E-17
EDA X 68835911 69259322 40 5.54E-05
ITIH6 X 54775332 54824673 4 5.67E-05
KLF8 X 56258822 56314322 2 2.53E-08
FAAH2 X 57313110 57515629 8 9.96E-13
ARHGEF9 X 62854848 63005426 7 1.11E-11
MTMR8 X 63487961 63615333 4 1.12E-06
ZC4H2 X 64136250 64254593 3 5.32E-10
VSIG4 X 65241580 65259967 3 1.09E-35
HEPH X 65382433 65487231 5 1.16E-96
EDA2R X 65815479 65859140 2 5.13E-190
AR X 66763874 66944119 6 2.02E-269
OPHN1 X 67262186 67653299 41 4.25E-190
STARD8 X 67867511 67945684 12 1.56E-17
EDA X 68835911 69259322 40 5.54E-05
Some discussion from the paper:
As mentioned above, the GWAS identified 247 independent autosomal loci and 40 independent X chromosome loci. The top 20 hits from the autosomes were located in/near to genes that have been associated with, for example, hair growth/length in mice (FGF5) [20], grey hair (IRF4) [21], cancer (breast: MEMO1 [22], bladder: SLC14A2 [23]), histone acetylation (HDAC9), and frontotemporal dementia (MAPT) [24]. Two of the top 10 X chromosome SNPs were located in OPHN1, a gene previously associated with X-linked mental retardation [25].
Of the top autosomal gene-based findings (maximum P=3.1x10^-15), RSPO2 has been linked to hair growth in dogs. PGDFA has been linked to hair follicle development [26]; EBF1 is expressed in dermal papillae in mature hair follicles [27]; PRR23B is proximal to a GWAS hit for eyebrow thickness [21]; and WNT10A has been linked to both straight hair [28] and dry hair [29].
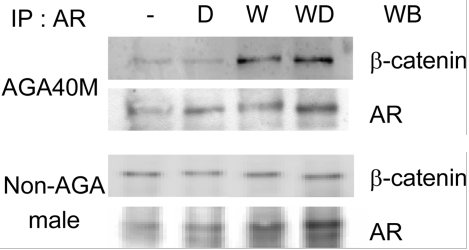
The top X chromosome gene-based findings included the androgen receptor (AR), which has been well established as a baldness associated gene [30], along with its upstream (EDA2R) and downstream (OPHN1) genes. EDA2R plays a role in the maintenance of hair and teeth as part of the tumor necrosis factor receptor. Onset of male pattern baldness could be influenced by EDA2R via activation of nuclear proto-oncoprotein c-Jun, which is linked to transcription activation of AR [31]. Two other genes included in the gene-based findings, OPHN1 and ZC4H2, have previously been associated with X-linked mental retardation [25, 32].
Seems simple, right?